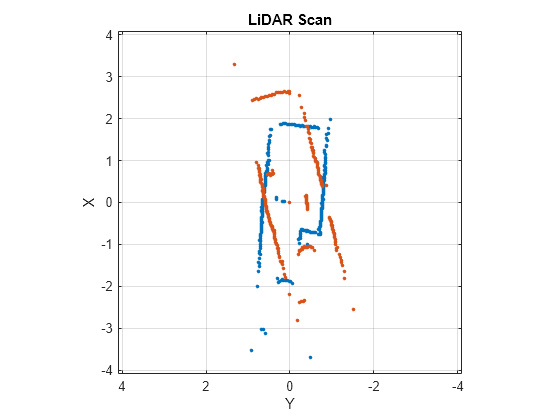
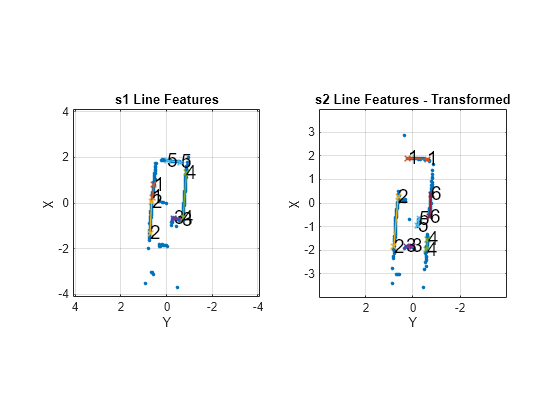
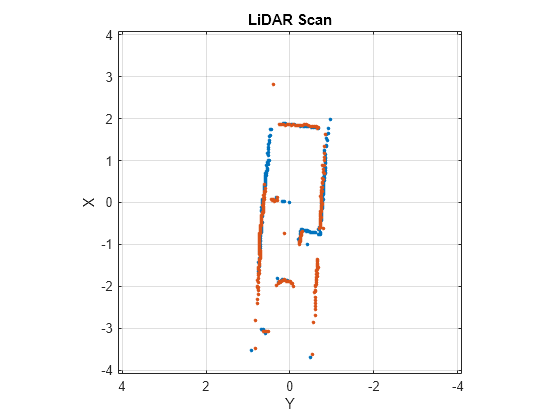
matchScansLine
Estimate pose between two laser scans using line features
Syntax
Description
relpose = matchScansLine(currScan,refScan,initialRelPose)initialRelPose.
[___] = matchScansLine(___,
specifies options using one or more name-value pair arguments.Name,Value)
Examples
Input Arguments
Name-Value Arguments
Output Arguments
References
[1] Neira, J., and J.d. Tardos. “Data Association in Stochastic Mapping Using the Joint Compatibility Test.” IEEE Transactions on Robotics and Automation 17, no. 6 (2001): 890–97. https://doi.org/10.1109/70.976019.
[2] Shen, Xiaotong, Emilio Frazzoli, Daniela Rus, and Marcelo H. Ang. “Fast Joint Compatibility Branch and Bound for Feature Cloud Matching.” 2016 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), 2016. https://doi.org/10.1109/iros.2016.7759281.
Version History
Introduced in R2020a