loss
Loss of linear incremental learning model on batch of data
Description
loss returns the regression or classification loss of a configured incremental learning model for linear regression (incrementalRegressionLinear object) or linear binary classification (incrementalClassificationLinear object).
To measure model performance on a data stream and store the results in the output model, call updateMetrics or updateMetricsAndFit.
Examples
The performance of an incremental model on streaming data is measured in three ways:
Cumulative metrics measure the performance since the start of incremental learning.
Window metrics measure the performance on a specified window of observations. The metrics are updated every time the model processes the specified window.
The
lossfunction measures the performance on a specified batch of data only.
Load the human activity data set. Randomly shuffle the data.
load humanactivity n = numel(actid); rng(1) % For reproducibility idx = randsample(n,n); X = feat(idx,:); Y = actid(idx);
For details on the data set, enter Description at the command line.
Responses can be one of five classes: Sitting, Standing, Walking, Running, or Dancing. Dichotomize the response by identifying whether the subject is moving (actid > 2).
Y = Y > 2;
Create an incremental linear SVM model for binary classification. Configure the model for loss by specifying the class names, prior class distribution (uniform), and arbitrary coefficient and bias values. Specify a metrics window size of 1000 observations.
p = size(X,2); Beta = randn(p,1); Bias = randn(1); Mdl = incrementalClassificationLinear('Beta',Beta,'Bias',Bias, ... 'ClassNames',unique(Y),'Prior','uniform','MetricsWindowSize',1000);
Mdl is an incrementalClassificationLinear model. All its properties are read-only. Instead of specifying arbitrary values, you can take either of these actions to configure the model:
Train an SVM model using
fitcsvmorfitclinearon a subset of the data (if available), and then convert the model to an incremental learner by usingincrementalLearner.Incrementally fit
Mdlto data by usingfit.
Simulate a data stream, and perform the following actions on each incoming chunk of 50 observations:
Call
updateMetricsto measure the cumulative performance and the performance within a window of observations. Overwrite the previous incremental model with a new one to track performance metrics.Call
lossto measure the model performance on the incoming chunk.Call
fitto fit the model to the incoming chunk. Overwrite the previous incremental model with a new one fitted to the incoming observations.Store all performance metrics to see how they evolve during incremental learning.
% Preallocation numObsPerChunk = 50; nchunk = floor(n/numObsPerChunk); ce = array2table(zeros(nchunk,3),'VariableNames',["Cumulative" "Window" "Loss"]); % Incremental learning for j = 1:nchunk ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); idx = ibegin:iend; Mdl = updateMetrics(Mdl,X(idx,:),Y(idx)); ce{j,["Cumulative" "Window"]} = Mdl.Metrics{"ClassificationError",:}; ce{j,"Loss"} = loss(Mdl,X(idx,:),Y(idx)); Mdl = fit(Mdl,X(idx,:),Y(idx)); end
Mdl is an incrementalClassificationLinear model object trained on all the data in the stream. During incremental learning and after the model is warmed up, updateMetrics checks the performance of the model on the incoming observations, then and the fit function fits the model to those observations. loss is agnostic of the metrics warm-up period, so it measures the classification error for all iterations.
To see how the performance metrics evolve during training, plot them.
figure plot(ce.Variables) xlim([0 nchunk]) ylim([0 0.05]) ylabel('Classification Error') xline(Mdl.MetricsWarmupPeriod/numObsPerChunk,'r-.') legend(ce.Properties.VariableNames) xlabel('Iteration')
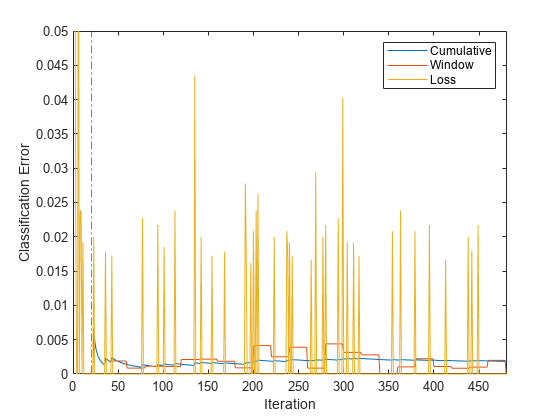
The yellow line represents the classification error on each incoming chunk of data. After the metrics warm-up period, Mdl tracks the cumulative and window metrics. The cumulative and batch losses converge as the fit function fits the incremental model to the incoming data.
Fit an incremental learning model for regression to streaming data, and compute the mean absolute deviation (MAD) on the incoming data batches.
Load the robot arm data set. Obtain the sample size n and the number of predictor variables p.
load robotarm
n = numel(ytrain);
p = size(Xtrain,2);For details on the data set, enter Description at the command line.
Create an incremental linear model for regression. Configure the model as follows:
Specify a metrics warm-up period of 1000 observations.
Specify a metrics window size of 500 observations.
Track the mean absolute deviation (MAD) to measure the performance of the model. Create an anonymous function that measures the absolute error of each new observation. Create a structure array containing the name
MeanAbsoluteErrorand its corresponding function.Configure the model to predict responses by specifying that all regression coefficients and the bias are 0.
maefcn = @(z,zfit,w)(abs(z - zfit)); maemetric = struct("MeanAbsoluteError",maefcn); Mdl = incrementalRegressionLinear('MetricsWarmupPeriod',1000,'MetricsWindowSize',500, ... 'Metrics',maemetric,'Beta',zeros(p,1),'Bias',0,'EstimationPeriod',0)
Mdl =
incrementalRegressionLinear
IsWarm: 0
Metrics: [2×2 table]
ResponseTransform: 'none'
Beta: [32×1 double]
Bias: 0
Learner: 'svm'
Properties, Methods
Mdl is an incrementalRegressionLinear model object configured for incremental learning.
Perform incremental learning. At each iteration:
Simulate a data stream by processing a chunk of 50 observations.
Call
updateMetricsto compute cumulative and window metrics on the incoming chunk of data. Overwrite the previous incremental model with a new one fitted to overwrite the previous metrics.Call
lossto compute the MAD on the incoming chunk of data. Whereas the cumulative and window metrics require that custom losses return the loss for each observation,lossrequires the loss on the entire chunk. Compute the mean of the absolute deviation.Call
fitto fit the incremental model to the incoming chunk of data.Store the cumulative, window, and chunk metrics to see how they evolve during incremental learning.
% Preallocation numObsPerChunk = 50; nchunk = floor(n/numObsPerChunk); mae = array2table(zeros(nchunk,3),'VariableNames',["Cumulative" "Window" "Chunk"]); % Incremental fitting for j = 1:nchunk ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); idx = ibegin:iend; Mdl = updateMetrics(Mdl,Xtrain(idx,:),ytrain(idx)); mae{j,1:2} = Mdl.Metrics{"MeanAbsoluteError",:}; mae{j,3} = loss(Mdl,Xtrain(idx,:),ytrain(idx),'LossFun',@(x,y,w)mean(maefcn(x,y,w))); Mdl = fit(Mdl,Xtrain(idx,:),ytrain(idx)); end
Mdl is an incrementalRegressionLinear model object trained on all the data in the stream. During incremental learning and after the model is warmed up, updateMetrics checks the performance of the model on the incoming observations, and the fit function fits the model to those observations.
Plot the performance metrics to see how they evolved during incremental learning.
figure h = plot(mae.Variables); xlim([0 nchunk]) ylabel('Mean Absolute Deviation') xline(Mdl.MetricsWarmupPeriod/numObsPerChunk,'r-.') xlabel('Iteration') legend(h,mae.Properties.VariableNames)
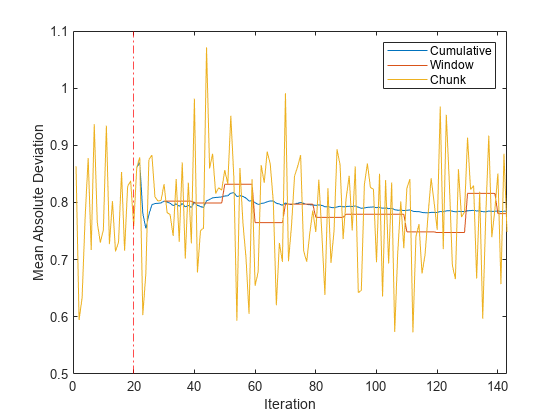
The plot suggests the following:
updateMetricscomputes the performance metrics after the metrics warm-up period only.updateMetricscomputes the cumulative metrics during each iteration.updateMetricscomputes the window metrics after processing 500 observations.Because
Mdlwas configured to predict observations from the beginning of incremental learning,losscan compute the MAD on each incoming chunk of data.
Input Arguments
Incremental learning model, specified as an incrementalClassificationLinear or incrementalRegressionLinear model object. You can create Mdl directly or by converting a supported, traditionally trained machine learning model using the incrementalLearner function. For more details, see the corresponding reference page.
You must configure Mdl to compute its loss on a batch of observations.
If
Mdlis a converted, traditionally trained model, you can compute its loss without any modifications.Otherwise,
Mdlmust satisfy the following criteria, which you can specify directly or by fittingMdlto data usingfitorupdateMetricsAndFit.If
Mdlis anincrementalRegressionLinearmodel, its model coefficientsMdl.Betaand biasMdl.Biasmust be nonempty arrays.If
Mdlis anincrementalClassificationLinearmodel, its model coefficientsMdl.Betaand biasMdl.Biasmust be nonempty arrays, the class namesMdl.ClassNamesmust contain two classes, and the prior class distributionMdl.Priormust contain known values.Regardless of object type, if you configure the model so that functions standardize predictor data, the predictor means
Mdl.Muand standard deviationsMdl.Sigmamust be nonempty arrays.
Batch of predictor data with which to compute the loss, specified as a
floating-point matrix of n observations and
Mdl.NumPredictors predictor variables. The value of the ObservationsIn name-value
argument determines the orientation of the variables and observations. The default
ObservationsIn value is "rows", which indicates that
observations in the predictor data are oriented along the rows of
X.
The length of the observation labels Y and the number of observations in X must be equal; Y( is the label of observation j (row or column) in j)X.
Note
loss supports only floating-point
input predictor data. If your input data includes categorical data, you must prepare an encoded
version of the categorical data. Use dummyvar to convert each categorical variable
to a numeric matrix of dummy variables. Then, concatenate all dummy variable matrices and any
other numeric predictors. For more details, see Dummy Variables.
Data Types: single | double
Batch of responses (labels) with which to compute the loss, specified as a categorical, character, or string array, logical or floating-point vector, or cell array of character vectors for classification problems; or a floating-point vector for regression problems.
The length of the observation labels Y and the number of
observations in X must be equal;
Y( is the label of observation
j (row or column) in j)X.
For classification problems:
losssupports binary classification only.If
Ycontains a label that is not a member ofMdl.ClassNames,lossissues an error.The data type of
YandMdl.ClassNamesmust be the same.
Data Types: char | string | cell | categorical | logical | single | double
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: 'ObservationsIn','columns','Weights',W specifies that the columns of the predictor matrix correspond to observations, and the vector W contains observation weights to apply.
Loss function, specified as the comma-separated pair consisting of 'LossFun' and a built-in loss function name or function handle.
Classification problems: The following table lists the available loss functions when
Mdlis anincrementalClassificationLinearmodel. Specify one using its corresponding character vector or string scalar.Name Description "binodeviance"Binomial deviance "classiferror"(default)Misclassification rate in decimal "exponential"Exponential loss "hinge"Hinge loss "logit"Logistic loss "quadratic"Quadratic loss For more details, see Classification Loss.
Logistic regression learners return posterior probabilities as classification scores, but SVM learners do not (see
predict).To specify a custom loss function, use function handle notation. The function must have this form:
lossval = lossfcn(C,S,W)
The output argument
lossvalis an n-by-1 floating-point vector, wherelossval(is the classification loss of observationj)jYou specify the function name (
lossfcnCis an n-by-2 logical matrix with rows indicating the class to which the corresponding observation belongs. The column order corresponds to the class order in theClassNamesproperty. CreateCby settingC(=p,q)1, if observationpqp0.Sis an n-by-2 numeric matrix of predicted classification scores.Sis similar to thescoreoutput ofpredict, where rows correspond to observations in the data and the column order corresponds to the class order in theClassNamesproperty.S(is the classification score of observationp,q)pqWis an n-by-1 numeric vector of observation weights.
Regression problems: The following table lists the available loss functions when
Mdlis anincrementalRegressionLinearmodel. Specify one using its corresponding character vector or string scalar.Name Description Learner Supporting Metric "epsiloninsensitive"Epsilon insensitive loss 'svm'"mse"(default)Weighted mean squared error 'svm'and'leastsquares'For more details, see Regression Loss.
To specify a custom loss function, use function handle notation. The function must have this form:
lossval = lossfcn(Y,YFit,W)
The output argument
lossvalis a floating-point scalar.You specify the function name (
lossfcnYis a length n numeric vector of observed responses.YFitis a length n numeric vector of corresponding predicted responses.Wis an n-by-1 numeric vector of observation weights.
Example: 'LossFun',"mse"
Example: 'LossFun',@lossfcn
Data Types: char | string | function_handle
Predictor data observation dimension, specified as the comma-separated pair consisting of 'ObservationsIn' and 'columns' or 'rows'.
Data Types: char | string
Batch of observation weights, specified as the comma-separated pair consisting of 'Weights' and a floating-point vector of positive values. loss weighs the observations in the input data with the corresponding values in Weights. The size of Weights must equal n, which is the number of observations in the input data.
By default, Weights is ones(.n,1)
For more details, see Observation Weights.
Data Types: double | single
Output Arguments
More About
Classification loss functions measure the predictive inaccuracy of classification models. When you compare the same type of loss among many models, a lower loss indicates a better predictive model.
Consider the following scenario.
L is the weighted average classification loss.
n is the sample size.
yj is the observed class label. The software codes it as –1 or 1, indicating the negative or positive class (or the first or second class in the
ClassNamesproperty), respectively.f(Xj) is the positive-class classification score for observation (row) j of the predictor data X.
mj = yjf(Xj) is the classification score for classifying observation j into the class corresponding to yj. Positive values of mj indicate correct classification and do not contribute much to the average loss. Negative values of mj indicate incorrect classification and contribute significantly to the average loss.
The weight for observation j is wj.
Given this scenario, the following table describes the supported loss functions that you
can specify by using the LossFun name-value argument.
| Loss Function | Value of LossFun | Equation |
|---|---|---|
| Binomial deviance | "binodeviance" | |
| Exponential loss | "exponential" | |
| Misclassification rate in decimal | "classiferror" | where is the class label corresponding to the class with the maximal score, and I{·} is the indicator function. |
| Hinge loss | "hinge" | |
| Logistic loss | "logit" | |
| Quadratic loss | "quadratic" |
The loss function does not omit an observation with a
NaN score when computing the weighted average loss. Therefore,
loss can return NaN when the predictor
data X contains missing values, and the name-value argument
LossFun is not specified as "classiferror". In
most cases, if the data set does not contain missing predictors, the
loss function does not return NaN.
This figure compares the loss functions over the score m for one observation. Some functions are normalized to pass through the point (0,1).

Regression loss functions measure the predictive inaccuracy of regression models. When you compare the same type of loss among many models, a lower loss indicates a better predictive model.
Consider the following scenario.
L is the weighted average classification loss.
n is the sample size.
yj is the observed response of observation j.
f(Xj) is the predicted value of observation j of the predictor data X.
The weight for observation j is wj.
Given this scenario, the following table describes the supported loss functions that you can specify by using the LossFun name-value argument.
| Loss Function | Value of LossFun | Equation |
|---|---|---|
| Epsilon insensitive loss | "epsiloninsensitive" | |
| Mean squared error | "mse" |
The loss function does not omit an observation with a
NaN prediction when computing the weighted average loss. Therefore,
loss can return NaN when the predictor
data X contains missing values. In most cases, if the data set does not
contain missing predictors, the loss function does not return
NaN.
Algorithms
For classification problems, if the prior class probability distribution is known (in other words, the prior distribution is not empirical), loss normalizes observation weights to sum to the prior class probabilities in the respective classes. This action implies that observation weights are the respective prior class probabilities by default.
For regression problems or if the prior class probability distribution is empirical, the software normalizes the specified observation weights to sum to 1 each time you call loss.
Extended Capabilities
Usage notes and limitations:
Use
saveLearnerForCoder,loadLearnerForCoder, andcodegen(MATLAB Coder) to generate code for thelossfunction. Save a trained model by usingsaveLearnerForCoder. Define an entry-point function that loads the saved model by usingloadLearnerForCoderand calls thelossfunction. Then usecodegento generate code for the entry-point function.To generate single-precision C/C++ code for
loss, specifyDataType="single"when you call theloadLearnerForCoderfunction.This table contains notes about the arguments of
loss. Arguments not included in this table are fully supported.Argument Notes and Limitations MdlFor usage notes and limitations of the model object, see
incrementalClassificationLinearorincrementalRegressionLinear.XBatch-to-batch, the number of observations can be a variable size, but must equal the number of observations in
Y.The number of predictor variables must equal to
Mdl.NumPredictors.Xmust besingleordouble.
YBatch-to-batch, the number of observations can be a variable size, but must equal the number of observations in
X.For classification problems, all labels in
Ymust be represented inMdl.ClassNames.YandMdl.ClassNamesmust have the same data type.
'LossFun'The specified function cannot be an anonymous function. If you configure
Mdlto shuffle data (Mdl.Shuffleistrue, orMdl.Solveris'sgd'or'asgd'), thelossfunction randomly shuffles each incoming batch of observations before it fits the model to the batch. The order of the shuffled observations might not match the order generated by MATLAB®. Therefore, if you fitMdlbefore computing the loss, the loss computed in MATLAB and those computed by the generated code might not be equal.Use a homogeneous data type for all floating-point input arguments and object properties, specifically, either
singleordouble.
For more information, see Introduction to Code Generation.
Version History
Introduced in R2020bThe loss function no longer omits an observation with a
NaN prediction (score for classification and response for regression)
when computing the weighted average loss. Therefore, loss can
now return NaN when the predictor data X contains
missing values, and the name-value argument LossFun is not specified as
"classiferror" (for classification). In most cases, if the data set
does not contain missing predictors, the loss function does not
return NaN.
If loss in your code returns NaN, you can
update your code to avoid this result. Remove or replace the missing values by using
rmmissing or fillmissing, respectively.
See Also
Objects
Functions
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)