phased.CFARDetector2D
Two-dimensional CFAR detector
Description
phased.CFARDetector2D
System object™ implements a constant false-alarm rate detector (CFAR) for selected
elements (called cells) of two-dimensional image data. A detection is declared when an
image cell value exceeds a threshold. To maintain a constant false alarm-rate, the
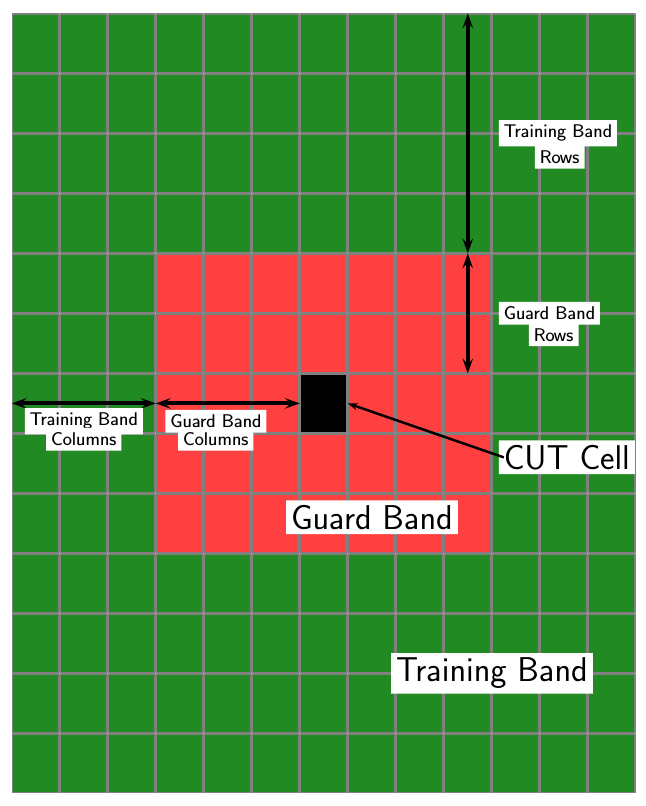
threshold is set to a multiple of the image noise power. The detector estimates noise
power for a cell-under-test (CUT) from surrounding cells using
one of three cell averaging methods, or an order statistics method. The cell-averaging
methods are cell averaging (CA), greatest-of cell averaging (GOCA), or smallest-of cell
averaging (SOCA).
For each test cell, the detector:
Estimates the noise statistic from the cell values in the training band surrounding the CUT cell.
Computes the threshold by multiplying the noise estimate by the threshold factor.
Compares the CUT cell value to the threshold to determine whether a target is present or absent. If the value is greater than the threshold, a target is present.
detector = phased.CFARDetector2Ddetector.
To run the 2-D CFAR detector:
Create the
phased.CFARDetector2Dobject and set its properties.Call the object with arguments, as if it were a function.
To learn more about how System objects work, see What Are System Objects?
Creation
Description
detector = phased.CFARDetector2D creates a 2-D CFAR
detector object, detector.
detector = phased.CFARDetector2D(Name,Value)detector, with each specified property
Name set to the specified Value.
You can specify additional name-value pair arguments in any order as
(Name1,Value1,...,NameN,ValueN).
Properties
Usage
Description
[Y,th] = detector(___) also returns the
detection threshold, th, applied to detected cells
under test. To enable this syntax, set the
ThresholdOutputPort property to
true.
Input Arguments
Output Arguments
Object Functions
To use an object function, specify the
System object as the first input argument. For
example, to release system resources of a System object named obj, use
this syntax:
release(obj)
Examples
More About
Algorithms
References
[1] Mott, H. Antennas for Radar and Communications. New York: John Wiley & Sons, 1992.
[2] Richards, M. A. Fundamentals of Radar Signal Processing. New York: McGraw-Hill, 2005.
[3] Skolnik, M. Introduction to Radar Systems, 3rd Ed. New York: McGraw-Hill, 2001.
Extended Capabilities
Version History
Introduced in R2016b