2-D CFAR Detector
Two-dimensional constant false alarm rate (CFAR) detector
Libraries:
Phased Array System Toolbox /
Detection
Description
The 2-D CFAR Detector block implements a constant false-alarm rate detector for two dimensional image data. A detection is declared when an image cell value exceeds a threshold. To maintain a constant false alarm-rate, the threshold is set to a multiple of the image noise power. The detector estimates noise power from neighboring cells surrounding the cell-under-test (CUT) using one of three cell averaging methods, or an order statistics method. The cell-averaging methods are cell-averaging (CA), greatest-of cell averaging (GOCA), or smallest-of cell averaging (SOCA).
For each test cell, the detector:
estimates the noise statistic from the cell values in the training band surrounding the CUT cell.
computes the threshold by multiplying the noise estimate by the threshold factor.
compares the CUT cell value to the threshold to determine whether a target is present or absent. If the value is greater than the threshold, a target is present.
Ports
Input
Output
Parameters
Algorithms
CFAR 2-D requires an estimate of the noise power. Noise power is computed from cells that are assumed not to contain any target signal. These cells are the training cells. Training cells form a band around the cell-under-test (CUT) cell but may be separated from the CUT cell by a guard band. The detection threshold is computed by multiplying the noise power by the threshold factor.
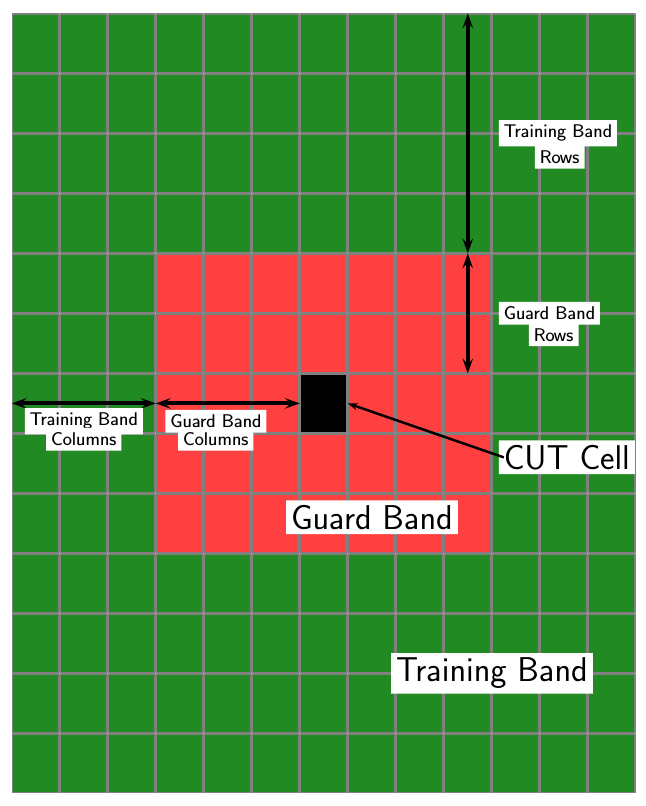
For GOCA and SOCA averaging, the noise power is derived from the mean value of one of the left or right halves of the training cell region.
Because the number of columns in the training region is odd, the cells in the middle column are assigned equally to either the left or right half.
When using the order-statistic method, the rank cannot be larger than the number of cells in the training cell region, Ntrain. You can compute Ntrain.
NTC is the number of training band columns.
NTR is the number of training band rows.
NGC is the number of guard band columns.
NGR is the number of guard band rows.
The total number of cells in the combined training region, guard region, and CUT cell is Ntotal = (2NTC + 2NGC + 1)(2NTR+ 2NGR + 1).
The total number of cells in the combined guard region and CUT cell is Nguard = (2NGC + 1)(2NGR + 1).
The number of training cells is Ntrain = Ntotal – Nguard.
By construction, the number of training cells is always even. Therefore, to implement a median filter, you can choose a rank of Ntrain/2 or Ntrain/2 + 1.
Version History
Introduced in R2016b