mergeDetections
Syntax
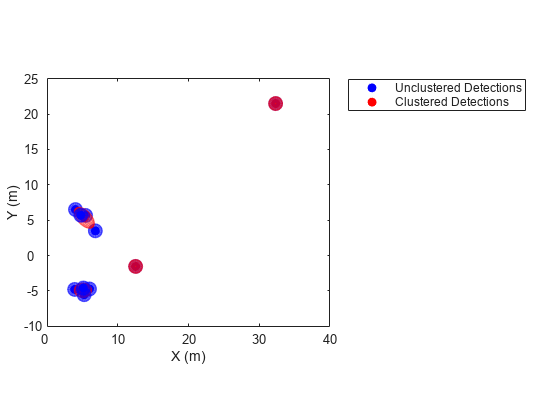
Description
clusteredDetections = mergeDetections(detections,clusterIndex)Time,
SensorIndex, ObjectClassID,
MeasurementParameters, and ObjectAttributes
properties or fields.
clusteredDetections = mergeDetections(___,MergingFcn=mergeFcn)