Get Started with Medical Image Labeler
The Medical Image
Labeler app enables you to explore and interactively label pixels in 2-D and 3-D
medical images. You can export labeled data as a groundTruthMedical object to train semantic segmentation algorithms. You can
publish snapshot images and animations with or without labels.

This tutorial provides an overview of the capabilities of the Medical Image Labeler and compares 2-D and 3-D image labeling. The typical app workflow includes these steps:
Open Medical Image Labeler App
Open the Medical Image Labeler app from the Apps tab
on the MATLAB® Toolstrip, under Image Processing and Computer
Vision. You can also load the app by using the
medicalImageLabeler command.
Create or Open Labeling Session
Manage labeling in the Medical Image Labeler using app sessions. Within one app session, you can import and label multiple image files. These might be repeat scans from one patient or scans from multiple patients with the same set of tissues, organs, or other regions of interest to label. The app enables you to create either a volume session or an image session. Use a volume session to label 3-D medical image data. Use an image session to label 2-D images or an image series, such as an ultrasound video.
You can either create a new labeling session or reopen a previous session:
New session — On the app toolstrip, click New Session and select New Volume session (3-D) or New Image session (2-D). In the dialog box that opens, specify a session folder. As you draw the label images, the app automatically saves them to the session folder. Therefore, the session folder must have access to enough memory to save all label images for the session. For more details, see the How Medical Image Labeler Manages Ground Truth Labels section.
Open session — On the app toolstrip, click Open Session and select one of the listed recent sessions, or click Open Session and navigate to a previous session folder.

Load Image Data
Load images to label from a file, from a medicalVolume or
medicalImage object in the workspace, or from a
groundTruthMedical object. Load image files when starting a new
labeling project. Load a medicalVolume or medicalImage
object when you have already loaded a file into the workspace, such as to perform
preprocessing and registration. Use groundTruthMedical objects when
working with labeled or partially labeled data from outside of MATLAB, or that have been shared from a different workstation. For more details
about working on a multi-person labeling team, see Collaborate on Multi-Labeler Medical Image Labeling Projects.
To load an image from a file, click Import and, under Data, select From File. The app supports loading these file formats, based on the type of labeling session:
Volume session — Single NIfTI, NRRD, DICOM, or TIFF file, or a directory containing multiple DICOM files corresponding to one image volume.
Image session — Single DICOM file, or any graphics file format listed in the
imformatsimage file format registry.
To load a medicalVolume or medicalImage object,
click Import, and, under Data, select
medicalVolume Object or medicalImage
Object, respectively. In the dialog box that opens, select the object to
import and click OK. You can only import one object at a
time.
To load a groundTruthMedical object, click
Import, and under Ground Truth, select
From File to load the object from a MAT file or From
Workspace to load the object from the MATLAB workspace. Importing a groundTruthMedical object loads
the image data, label data, and label definitions stored in the object into the
app.
The Data Browser pane lists all of the image files currently loaded in the app. Click a filename in the Data Browser to change the file to display and label.
Visually Explore Data
The app displays 3-D image data using individual panes for the Transverse, Sagittal, and Coronal slice planes and a 3-D Volume pane. For an example of how to customize the display of 3-D images, and publish images and animations, see Visualize 3-D Medical Image Data Using Medical Image Labeler. The app displays 2-D image data in the Slice pane. For details about navigating frames or adjusting the brightness and contrast of a 2-D image series, see Label 2-D Ultrasound Series Using Medical Image Labeler.

Create Label Definitions
A label definition specifies the name, color, and order of each label assigned to the image. You must use the same label definitions across all images within an app session. The Medical Image Labeler app supports pixel labeling.
You can create a label definition interactively in the app by clicking Create Label Definition in the Label Definitions pane. Optionally, you can assign a name for the label by clicking it, or change the color of the label by clicking the color square next to the label name.

Alternatively, import label definitions from a label definitions file or as part of a
groundTruthMedical object. You can create a label definitions file
programmatically, or load one exported from a previous app session. For more details
about creating a label definitions file programmatically, see the LabelDefinitions property of the groundTruthMedical
object.
Label Image
Label pixels using the tools in the Draw and Automate tabs of the app toolstrip. The app provides manual, semi-automated, and automated labeling tools.
Manual Labeling
Manually draw labels using the Freehand, Assisted Freehand, Polygon, and Paintbrush tools.
| Tool | Description | Image |
|---|---|---|
| Freehand | Draw a freehand shape. Click and drag to draw a line that follows the movement of the pointer. Click to add waypoints along the edge as you draw. Click and move the pointer without dragging to draw a straight line edge. Press Backspace to remove the most recently added waypoint and continue drawing. When you are finished, double-click or press Enter to close the freehand shape and stop drawing. |
|
| Assisted Freehand | Draw a freehand shape that snaps to the edges of the underlying image. Click to place the first waypoint. As you move the pointer, the line you draw automatically follows the edges in the underlying image. Click to add waypoints along the edge as you draw. To draw an unassisted edge that follows your pointer movement, click and drag between waypoints. Press Backspace to remove the most recently added waypoint and continue drawing. When you are finished, double-click or press Enter to close the freehand shape and stop drawing. |
|
| Polygon | Draw a polygon shape. Click to place each waypoint, which the tool connects with straight line edges. Press Backspace to remove the most recently added vertex and continue drawing. When you are finished, double-click or press Enter to close the polygon shape and stop drawing. |
|
| Paintbrush | Manually paint over the image to label pixels. Click and drag over the image to label pixels. Adjust the Brush Size to change the size of the paint brush. |
|
Once you click outside a newly drawn freehand, assisted freehand, or polygon region, the ROI boundary disappears. To manually refine the region, in the app toolstrip, click Select Drawn Region and click inside the label region. The label region becomes an ROI that you can edit. To move the ROI, click and drag on the boundary of the ROI between two waypoints. To change the shape of the ROI, click and drag a waypoint to move it, or right click and select Remove Waypoint to remove the waypoint. Right click inside the ROI to view options to cut, copy, delete, or reassign the region to a new label definition.

You can also manually refine labels by using the Eraser tool. Click and drag over labeled pixels to clear the label.
AI-Based Labeling
Medical Segment Anything Model (MedSAM) is a deep learning algorithm for the easy,
interactive segmentation of 2-D medical images and 2-D cross-sections of 3-D medial
volumes. You can use MedSAM to segment objects in several medical imaging
modalities, without needing to retrain the model for your data set. MedSAM builds
upon the foundation of the Segment Anything Model (SAM). Interactively segment
objects in medical images and in cross-sections of medical volumes using the MedSAM
algorithm in Medical Image Labeler. To get started, see Get Started with MedSAM in Medical Image Labeler. 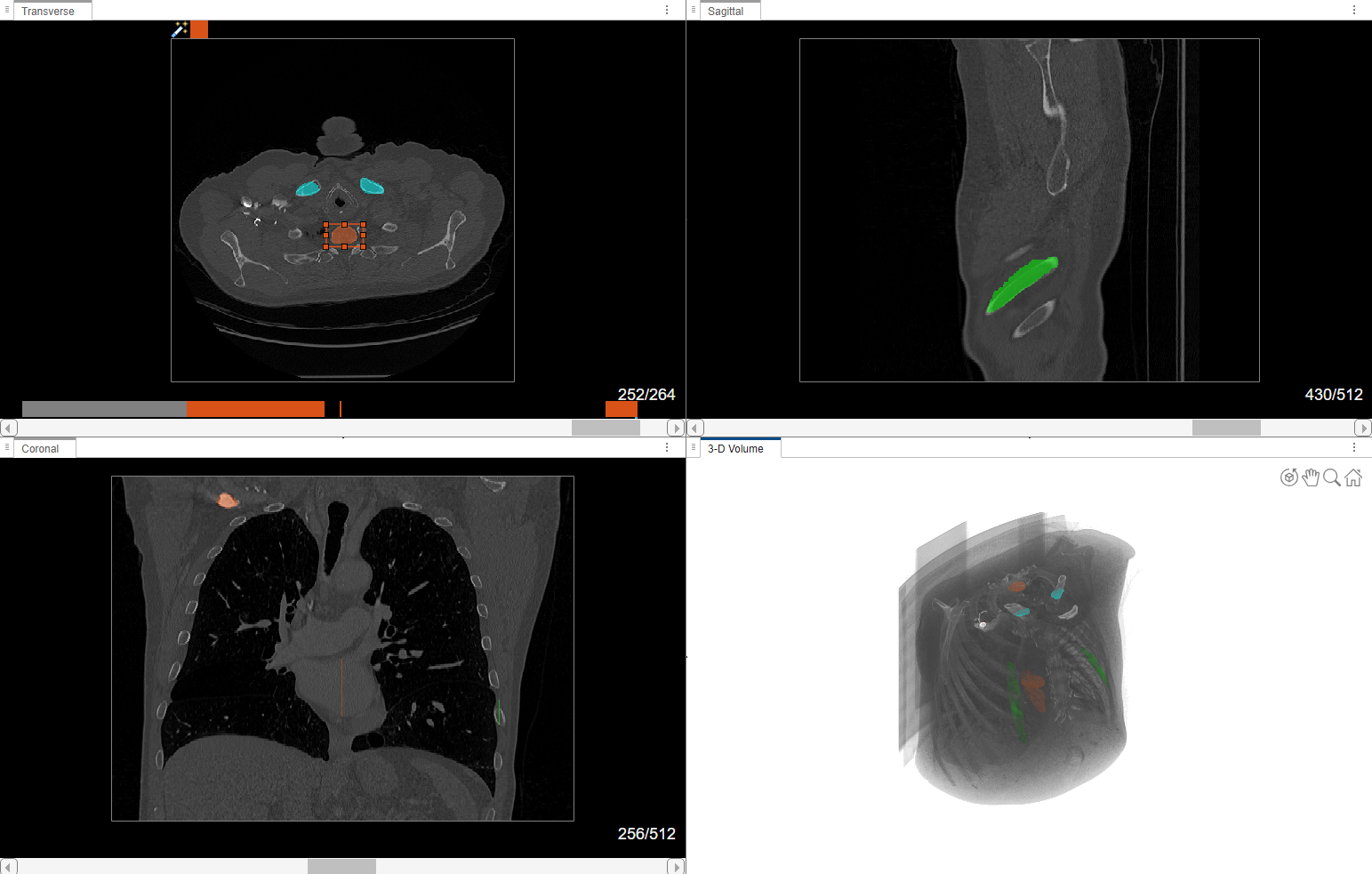
Semi-Automated Labeling
Add labels using semi-automated tools including Fill Region, Paint by Superpixels, and Trace Boundary. You can interpolate labeled regions between image frames or volume slices using the Auto Interpolate and Manually Interpolate tools.
| Tool | Description | Image |
|---|---|---|
| Fill Region | Flood fill label image or fill holes in label region. Select Fill Region in the app toolstrip, and click on the background or labeled region you want to flood fill. |
|
| Paint by Superpixels | Manually paint within an adjustable-sized grid of pixels. To use this tool, first select Paint Brush and then click Paint by Superpixels. Each superpixel contains a cluster of similar intensity values. Adjust the Superpixels Size to change the size of the superpixel grid. |
|
| Trace Boundary | Label connected regions that have similar intensity values. Select Trace Boundary in the app toolstrip, and then pause on a seed pixel or voxel in the region you want to label. The tool predicts the boundary of the region by including pixels or voxels with intensities that are similar to the current seed. Use the Threshold slider to adjust the similarity threshold used to predict the region boundary. Increasing the threshold includes a wider range of intensities above and below the seed value in the predicted region. Move the pointer to change the seed. |
|
Automated Labeling
The Automate tab contains built-in automation algorithms to refine existing labels or fully automate labeling. The app provides slice-based algorithms including Active Contours, Adaptive Threshold, Dilate, and Erode. In a volume session, the app additionally provides the Filter and Threshold, Smooth Edges, and Otsu's Threshold algorithms, which you can apply to all slices or to a specified slice range.
You can add a custom automation algorithm to use in the app. On the Automate tab, click Add Algorithm. Import an existing algorithm by selecting From File, or create a new algorithm using the provided function or class template. See Automate Labeling in Medical Image Labeler for an example that applies a custom automation algorithm for 2-D labeling.
MONAI Label
Medical Open Network for AI (MONAI) Label [1],[2] is a deep learning framework for labeling medical images. You can connect to MONAI Label within Medical Image Labeler to apply fully automated and interactive deep learning models for segmenting radiology images. To get started, see Get Started with MONAI Label in Medical Image Labeler.
These images show CT scans labeled using automated MONAI Label models in the Medical Image Labeler app.

This functionality requires the Medical Imaging Toolbox™ Interface for MONAI Label Library support package. You can install the support package from Add-On Explorer. For more information about installing add-ons, see Get and Manage Add-Ons.
Labeling Examples
For an example of labeling 2-D image data, see Label 2-D Ultrasound Series Using Medical Image Labeler.
For an example of labeling 3-D image data, see Label 3-D Medical Image Using Medical Image Labeler.
For an example of labeling using MONAI Label, see Segment CT Scan Using MONAI Label.
Export Labeling Results
You can export the groundTruthMedical object and label definitions as
files to share with a colleague.
To export the
groundTruthMedicalobject as a MAT file, on the Labeler tab, click Export and, under Ground Truth, select To File.To export the label definitions as a MAT file, on the Labeler tab, click Export and, under Label Definitions, select To File.
Note
The app stores the actual pixel label data in the LabelData
subfolder of the session folder. See How Medical Image Labeler Manages Ground Truth Labels for
details.
How Medical Image Labeler Manages Ground Truth Labels
As you label images in the Medical Image Labeler app, the app automatically saves three sets of data in the session folder associated with the current app session.
A
groundTruthMedicalobject stored as a MAT file. ThegroundTruthMedicalobject specifies the file locations of the unlabeled images and corresponding label images, as well as the name and color associated with each label.A subfolder named
LabelData, which contains the label images.The app saves pixel label images created in an image session in the
LabelDatafolder as MAT files. A MAT file stores the pixel labels as auint8array. You can read the label image into the MATLAB workspace by using theloadfunction.The app saves voxel label images created in a volume session in the
LabelDatafolder as NIfTI files or as MAT files that each contain a singlemedicalVolumeobject. The app saves labels as NIfTI files in most cases, and saves labels as a MAT file only when you load data from TIFF files ofmedicalVolumeobjects with incomplete spatial referencing. The NIfTI or MAT file stores the voxel labels as auint8array.A subfolder named
AppData, which contains data about the app session stored as a MAT file.
See Also
Medical Image
Labeler | groundTruthMedical





