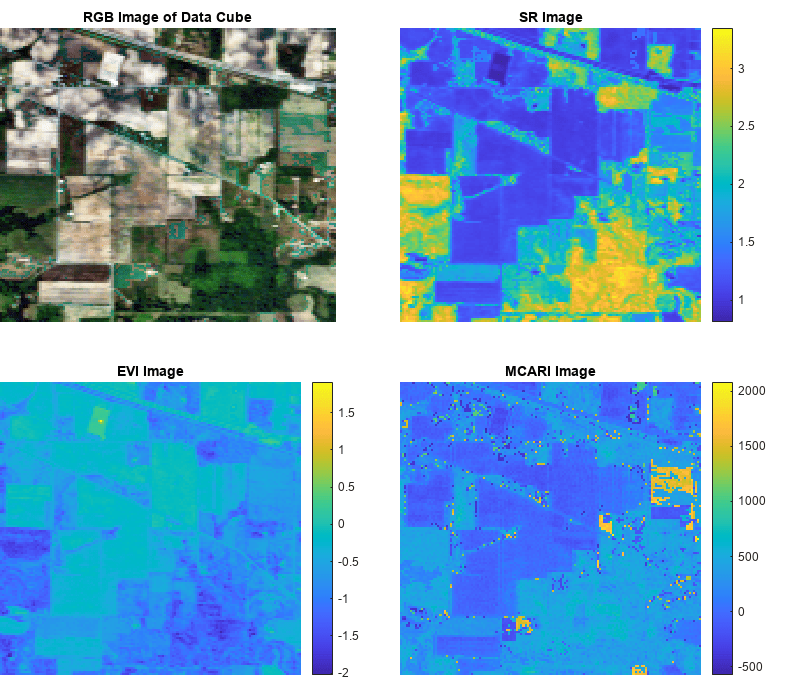
spectralIndices
Compute spectral indices
Syntax
Description
indices = spectralIndices(spcube)spcube to compute the greenness indices.
indices = spectralIndices(spcube,indexNames)indexNames.
indices = spectralIndices(___,BlockSize=blocksize)
The function divides the input image into distinct blocks,
processes each block, and then concatenates the processed output of each block to form the
output matrix. Spectral images are multi-dimensional data sets that can be too large to fit in
system memory in their entirety. This can cause the system to run out of memory while running
the spectralIndices function. If you encounter such an issue, perform block
processing by using this syntax.
For example, spectralIndices(spcube,BlockSize=[50 50]) divides the
input image into non-overlapping blocks of size 50-by-50 and then computes the spectral
indices for pixels in each block.
Note
This function requires the Hyperspectral Imaging Library for Image Processing Toolbox™. You can install the Hyperspectral Imaging Library for Image Processing Toolbox from Add-On Explorer. For more information about installing add-ons, see Get and Manage Add-Ons.
The Hyperspectral Imaging Library for Image Processing Toolbox requires desktop MATLAB®, as MATLAB Online™ and MATLAB Mobile™ do not support the library.