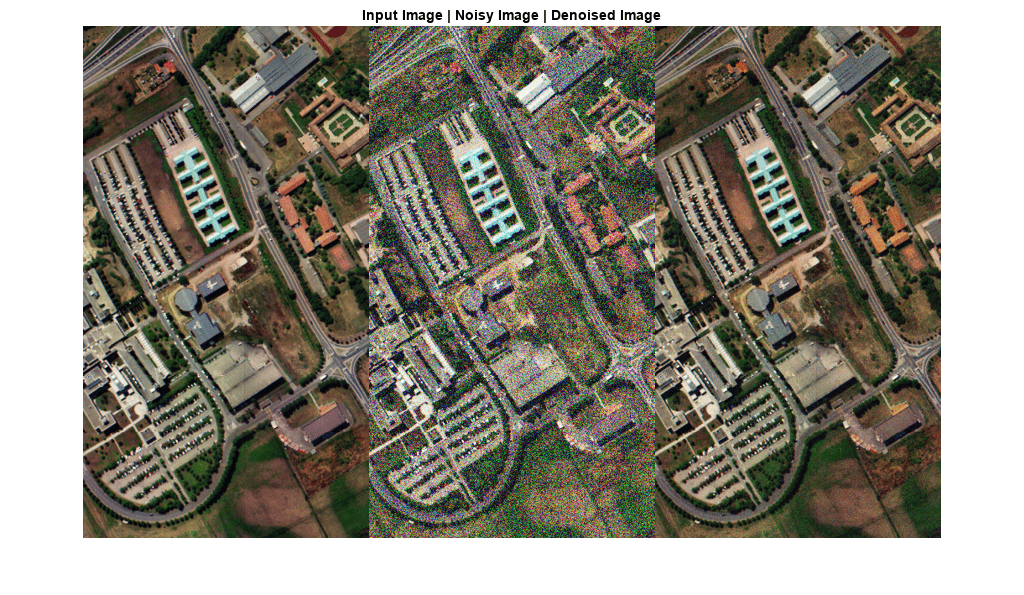
denoiseNGMeet
Denoise hyperspectral images using non-local meets global approach
Description
outputData = denoiseNGMeet(inputData)
outputData = denoiseNGMeet(inputData,Name=Value)
Note
This function requires the Hyperspectral Imaging Library for Image Processing Toolbox™. You can install the Hyperspectral Imaging Library for Image Processing Toolbox from Add-On Explorer. For more information about installing add-ons, see Get and Manage Add-Ons.
The Hyperspectral Imaging Library for Image Processing Toolbox requires desktop MATLAB®, as MATLAB Online™ and MATLAB Mobile™ do not support the library.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
Algorithms
The NGMeet method estimates the denoised data cube by using these steps. For each iteration, i
Compute spectral low-rank approximation of the noisy input data (Yi) by using singular value decomposition. The approximation results in a reduced data cube (Mi) and the related orthogonal basis Ai.
Perform spatial denoising of the reduced data cube Mi by using non-local similarity filtering. You can control the degree of smoothing by specifying the smoothing parameter
'Sigma'.Perform inverse projection. Map the denoised reduced data cube Mi to original space by using the orthogonal basis Ai. The result is the denoised output (Xi) obtained at iteration i.
Perform iterative regularization. Update the noisy input data, Yi+1 = λXi + (1-λ)Yi.
Repeat steps 1 to 4, for the specified number of iterations. The final value Xi is the denoised hyperspectral data.
References
[1] He, Wei, Quanming Yao, Chao Li, Naoto Yokoya, and Qibin Zhao. “Non-Local Meets Global: An Integrated Paradigm for Hyperspectral Denoising.” In 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 6861–70. Long Beach, CA, USA: IEEE, 2019. https://doi.org/10.1109/CVPR.2019.00703.