Update the Network Learnables for a Battery State of Charge Estimation Model
This example shows how to update the learnables of a deep learning model that estimates the state of charge while the Simulink model is simulating.
Battery state of charge (SOC) is the level of charge of that an electric battery has relative to its capacity, measured as a percentage. SOC is critical information for vehicle energy management systems and must be accurately estimated to ensure reliable electrified vehicles.
In this example, you:
Generate plain C++ code that does not depend on third party libraries for an LSTM network that predicts the battery SOC.
Run the generated code by using the Simulink external model on the host PC and update the network learnables during simulation.
Deploy the generated code on a Raspberry Pi board, copy the updated learnables from the host PC to the Raspberry Pi, and update the network learnables while the generated code is running.
Load Partially Trained LSTM Network
Load a partially trained LSTM network that predicts the battery SOC. This network has been trained using 10% of the data from the LG_HG2_Prepared_Dataset_McMasterUniversity_Jan_2020 data set. The model has these characteristics:
Because this example uses an LSTM network that can learn long-term trends, it uses only three features from the data set. The average voltage and average current are not required.
To learn long-term dependencies in the sequence data, the network contains two LSTM layers with 128 and 64 hidden units, respectively.
To reduce overfitting, the network contains two dropout layers with a dropout probability of
0.2.To match the size of the output, the network includes a fully connected layer.
To bound the output in the interval
[0,1], the network includes a sigmoid layer.
dlnet = load('dlnetSocEstimation.mat').dlnetSocEstimation;
display(dlnet.Layers); 7×1 Layer array with layers:
1 'sequenceinput' Sequence Input Sequence input with 3 dimensions
2 'lstm_1' LSTM LSTM with 128 hidden units
3 'dropout_1' Dropout 20% dropout
4 'lstm_2' LSTM LSTM with 64 hidden units
5 'dropout_2' Dropout 20% dropout
6 'fc' Fully Connected 1 fully connected layer
7 'layer' Sigmoid sigmoid
Check for Input Layer Normalization
The helper function checkForInputLayerNormalization checks whether the deep learning network has input layers where the setting of the Normalization property is not none. Because input layer normalization parameters are not Learnables, when the training data statistics change during retraining, the statement dlnet.Learnables = dlnetRetrained.Learnables does not guarantee that dlnet is equals to dlnetRetrained. Consider using normalization option none if the training data statistics change, otherwise, you can set the option ResetInputNormalization to false when using the trainnet function to keep the normalization parameters unchanged.
checkForInputLayerNormalization(dlnet);
Download Data Set
The downloadData helper function downloads the LG_HG2_Prepared_Dataset_McMasterUniversity_Jan_2020 data set and returns the training, validation, and testing data.
[XTrain, YTrain, XVal, YVal, XTest, YTest] = downloadData();
Downloading LGHG2@n10C_to_25degC.zip (56 MB) ...
Retrain LSTM Network
Retrain the LSTM network using the trainnet function on the entire data set to improve its accuracy. The getTrainingOptions helper function sets the training options for the network. Specify the loss function as mean-squared error to minimize the output root mean squared error (RMSE).
options = getTrainingOptions(XVal,YVal);
dlnet = trainnet(XTrain,YTrain,dlnet,"mean-squared-error",options);Save Updated Learnables
Save the updated learnables to a file by using the coder.write function.
updatedLearnablesValue = cellfun(@extractdata, dlnet.Learnables.Value, UniformOutput=false);
coder.write('learnablesValue.coderdata', updatedLearnablesValue);Wrote file 'learnablesValue.coderdata'. You can read this file with 'coder.read'.
Prepare Inputs for Simulink Model
Create a variable named Ts to control the step size. Create a timeseries object, Xin, from the predictors and Yin from the expected output.
Ts = 5e-4; stopTime = size(XTest,2)*Ts; timeVector = (0:Ts:(stopTime-Ts))'; Xin = timeseries(XTest', timeVector, Name='predictors'); Yin = timeseries(YTest', timeVector, Name='ground-truth');
Configure and Build the Simulink Model
Use this code to open the Simulink model and set the fixed-step size to Ts. Then, set the simulation mode to external and start the simulation.
modelName = 'deepLearningSocModel'; open_system(modelName); model = load_system(modelName); set_param(model, FixedStep='Ts'); set_param(model,SimulationMode='external'); set_param(model,SimulationCommand='start'); set_param(model,SimulationCommand='disconnect');
Update the Learnables of the Deployed Model Using Simulink External Mode
The Simulink model opens.
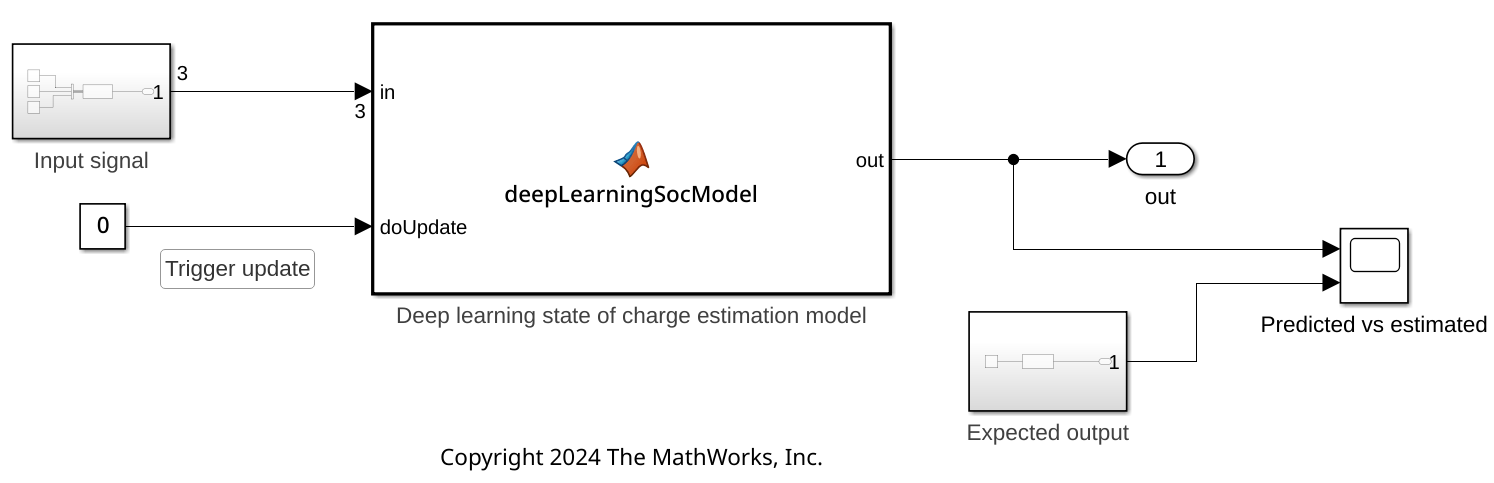
On the Hardware tab, use the Monitor & Tune button to run the system on the host by using Simulink external mode.
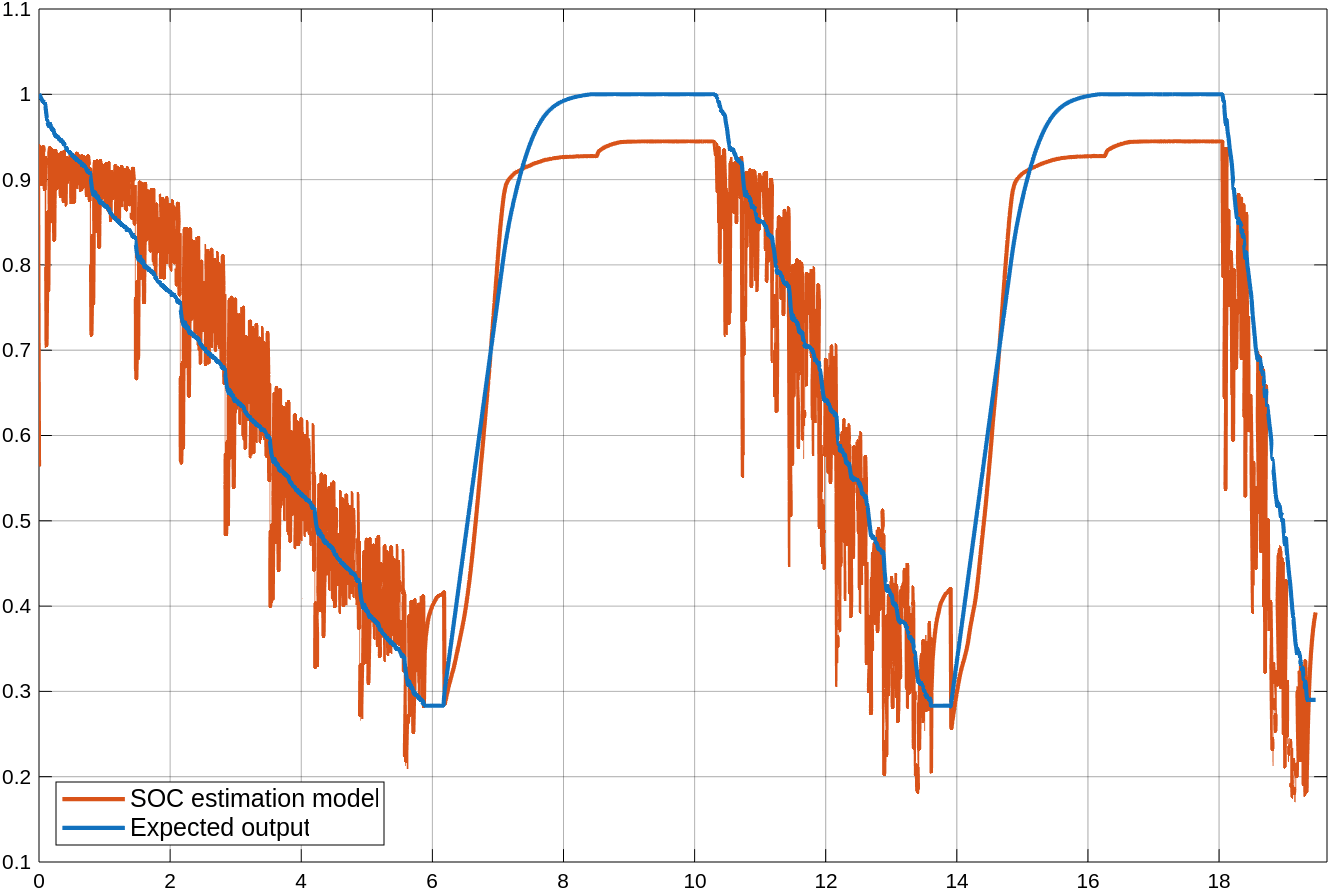
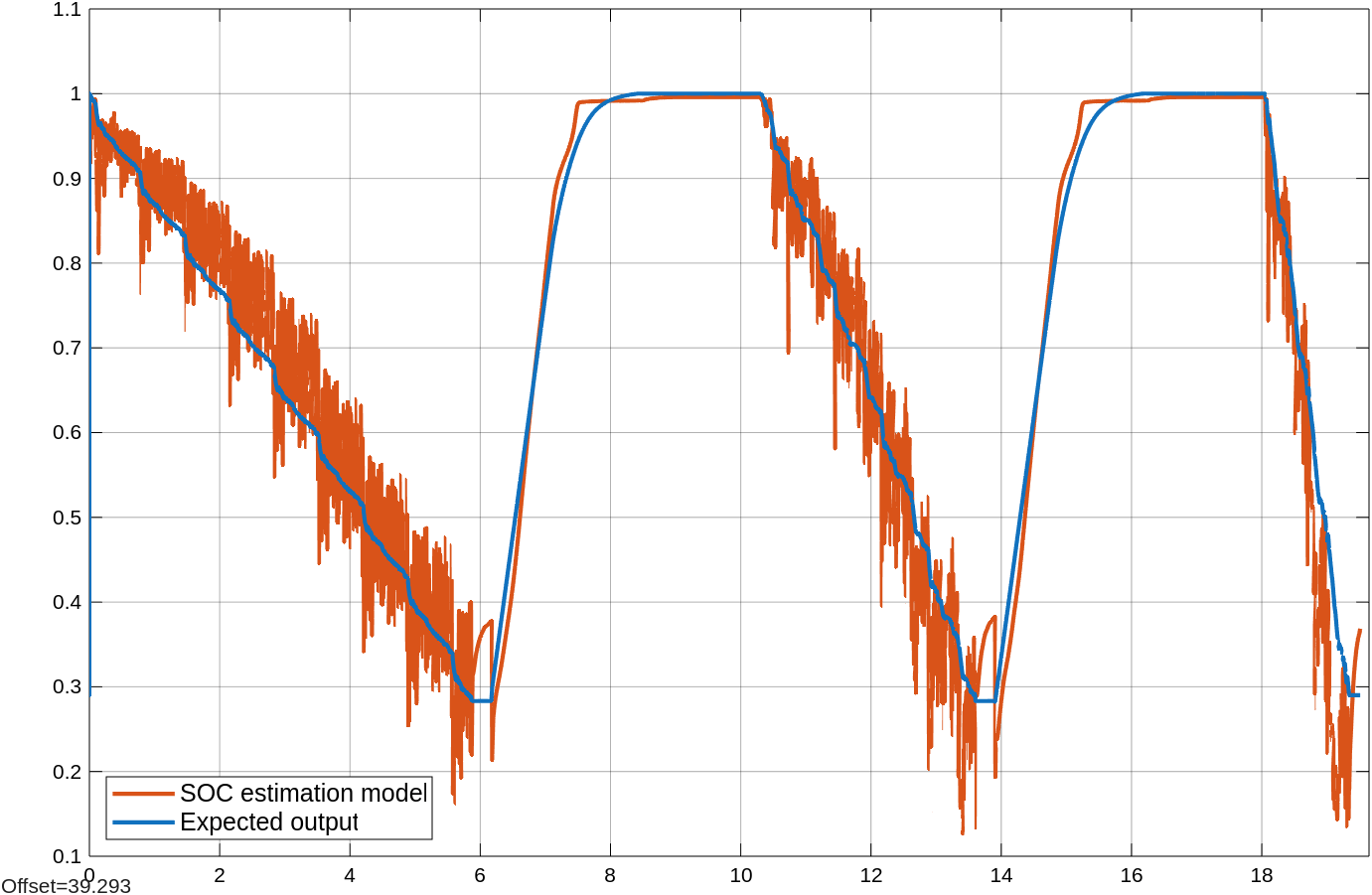
Open the Scope block named Predicted vs expected. The monitor displays the predictions of the partially trained LSTM network in red and the expected output in blue.
Click the Trigger update button to change the value of the doUpdate signal to 1 momentarily. It triggers the update of learnables of deep learning model in the MATLAB Function Block. The updated learnables is loaded while the model is simulating by using the coder.read function.
Inspect the Predicted vs expected Scope block once again. The model's predictions are more accurate and align more closely with the expected output.
 's
's
Deploy the Model on a Raspberry Pi Board and Run It Using External Mode
Note you need Embedded Coder® to deploy the model on a specific hardware. This example also requires Simulink Support Package for Raspberry Pi Hardware to deploy the model to a Raspberry Pi board. For more information, see Raspberry Pi Hardware.
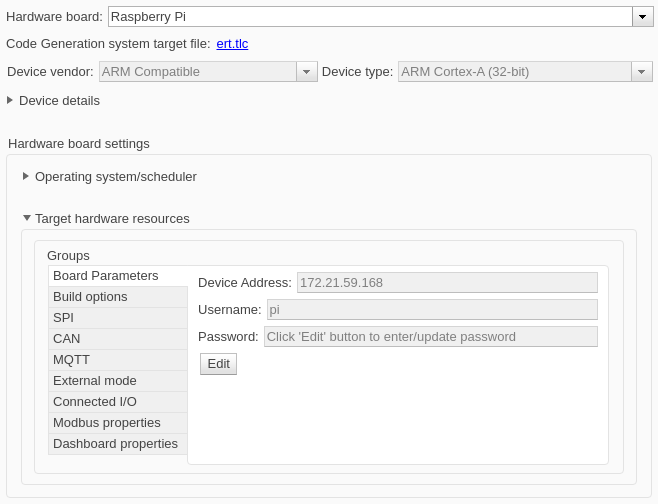
To run the model on a Raspberry Pi board, in Simulink Toolstrip, click Model Settings to open the Configuration Parameters dialog box. In the Hardware Implementation pane, set Hardware board to Raspberry Pi. Expand the Target hardware resources section and enter the Device Address, Username, and Password parameters for the board.
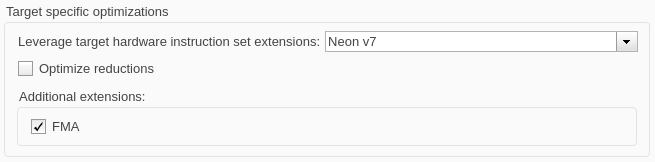
In the Code Generation > Optimization pane, in the Target specific optimizations section, set Leverage target hardware instruction set extensions to Neon v7. To optimize the performance of generated code, select the FMA box to leverage fast multiply-add.
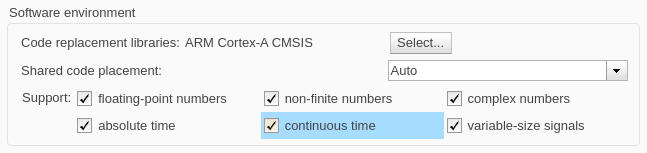
In the Code Generation > Interface pane, select the continuous time option to support the Repeating Sequence blocks in the Input signal and Expected output subsystems.
Click OK. In the Hardware tab, confirm the Hardware board is Raspberry Pi, and the Mode setting is Run on board. Click the Monitor & Tune button to run the model on the Raspberry Pi.

After the system starts to run, the Predicted vs. expected Scope block displays the same results as when you ran the model on the host computer.
Move Updated Learnables to the Raspberry Pi Board
Copy the learnables file to the Raspberry Pi. You can establish a connection with the board using the raspi command. Then, move the learnablesValue.coderdata file to the board by using the putFile function and close the connection.
r = raspi; putFile(r, 'learnablesValue.coderdata', '/home/username/learnablesValue.coderdata'); clear('r');
If this is your first time connecting to a Raspberry Pi board or you want to connect to a different board, use this line of code and replace username, password, and ipaddress with the actual values for the Raspberry Pi board.
r = raspi(iaddress, username, password);
After moving the coderdata file to the board, use the Trigger update button to update the learnables on the hardware. The scope displays the updated prediction results.
Helper Functions
checkForInputLayerNormalization
The checkForInputNormalization function throws a warning if the deep learning network has input layers where the setting of the Normalization property is not none.
function checkForInputLayerNormalization(dlnet) inputNames = dlnet.InputNames; for i = 1:numel(inputNames) inputLayer = getLayer(dlnet, inputNames{i}); if isprop(inputLayer, 'Normalization') if ~strcmpi(inputLayer.Normalization, 'none') warning("Input layer '" + inputLayer(1).Name + "' uses normalization option '" + inputLayer(1).Normalization + ... "'. Consider using normalization option 'none' if the training data statistics change. Otherwise, consider setting the option " + ... "'ResetInputNormalization' to 'false' when retraining the network to keep the normalization parameters unchanged. Input layer normalization parameters are not Learnables, " + ... "and therefore, when the training data statistics change during retraining, the statement dlnet.Learnables = dlnetRetrained.Learnables does not guarantee that dlnet will be equal " + ... "to dlnetRetrained."); end end end end
downloadData
The downloadData function downloads the LG_HG2_Prepared_Dataset_McMasterUniversity_Jan_2020 data set and returns the training, validation, and testing data. This function sets the subsequence length to 500 and the number of input features to 3.
function [XTrain, YTrain, XVal, YVal, XTest, YTest] = downloadData() url = "https://data.mendeley.com/public-files/datasets/cp3473x7xv/files/ad7ac5c9-2b9e-458a-a91f-6f3da449bdfb/file_downloaded"; downloadFolder = tempdir; outputFolder = fullfile(downloadFolder,"LGHG2@n10C_to_25degC"); if ~isfolder(outputFolder + "/dir") fprintf("Downloading LGHG2@n10C_to_25degC.zip (56 MB) ... ") filename = fullfile(downloadFolder,"LGHG2@n10C_to_25degC.zip"); websave(filename,url); unzip(filename,outputFolder) end chunkSize = 500; numFeatures = 3; trainingFile = fullfile(outputFolder,"Train","TRAIN_LGHG2@n10degC_to_25degC_Norm_5Inputs.mat"); [XTrain,YTrain] = setupData(trainingFile,chunkSize,numFeatures); validationFile = fullfile(outputFolder,"Validation", "01_TEST_LGHG2@n10degC_Norm_(05_Inputs).mat"); [XVal,YVal] = setupData(validationFile,chunkSize,numFeatures); testFile = fullfile(outputFolder,"Test","01_TEST_LGHG2@n10degC_Norm_(05_Inputs).mat"); S = load(testFile); XTest = S.X(1:numFeatures,:); YTest = S.Y; end
setupData
The setupData function loads a structure stored in the MAT file filename, extracts the first numFeatures features of the sequence data and the target values, and splits the data into subsequences of length chunkSize.
function [X,Y] = setupData(filename,chunkSize,numFeatures) S = load(filename); nSamples = length(S.Y); nElems = floor(nSamples/chunkSize); X = cell(nElems,1); Y = cell(nElems,1); for ii = 1:nElems idxStart = 1+(ii-1)*chunkSize; idxEnd = ii*chunkSize; X{ii} = S.X(1:numFeatures,idxStart:idxEnd); Y{ii} = S.Y(idxStart:idxEnd); end end
getTrainingOptions
The getTrainingOptions sets these training options:
Train for 100 epochs with mini-batches of size 64 using the
"adam"solver.Set initial learning rate to 0.01, a learning rate drop period of 45 and a learning rate drop factor of 0.1.
Set the gradient threshold to 1.
Shuffle the training data every epoch.
Specify the validation data.
Specify the input and target data formats as
CTB(channel, time, batch).Return the network with the lowest validation loss.
Display the training progress and suppress the verbose output.
function options = getTrainingOptions(XVal,YVal) options = trainingOptions("adam", ... MiniBatchSize = 64, ... MaxEpochs = 45, ... InitialLearnRate = 1e-2, ... LearnRateSchedule = "piecewise", ... LearnRateDropPeriod = 30, ... LearnRateDropFactor = 0.1, ... GradientThreshold = 1, ... Shuffle = "every-epoch", ... ValidationData = {XVal,YVal}, ... ValidationFrequency = 50, ... InputDataFormats="CTB", ... TargetDataFormats="CTB", ... OutputNetwork="best-validation-loss", ... Plots = "none", ... Verbose = false); end
See Also
Topics
- Generate Code for Battery State of Charge Estimation Using Deep Learning (Deep Learning Toolbox)