Convert Image Labeler Polygons to Labeled Blocked Image for Semantic Segmentation
This example shows how to convert polygon labels stored in a groundTruth object into a labeled blocked image suitable for semantic segmentation workflows.
You can use the Image Labeler app in Computer Vision Toolbox to label images that are too large to fit into memory and multiresolution images. For more information, see Label Large Images in the Image Labeler (Computer Vision Toolbox). The Image Labeler app does not support pixel labeling of blocked images. You can only create labels using ROI shapes such as polygons, rectangles, and lines. This example shows how you can use the polyToBlockedImage function to convert polygon ROIs into a pixel-labeled blocked image for semantic segmentation workflows.
Create a blocked image using a modified version of a training image of a lymph node containing tumor tissue, from the CAMELYON16 data set. The modified image has three coarse resolution levels. The spatial referencing has been adjusted to enforce a consistent aspect ratio and to register features at each level.
bim = blockedImage("tumor_091R.tif");Load Label Ground Truth Data
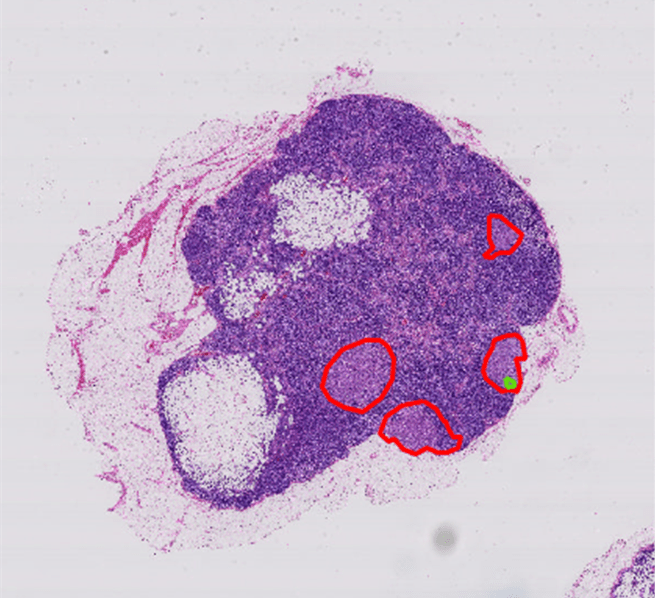
This example loads a presaved groundTruth object, gTruth.mat, created by labeling the blocked image data in bim using the Image Labeler app. The groundTruth object stores the polygon labels displayed in the figure. A normal tissue ROI is outlined in green, and tumor tissue ROIs are outlined in red. You can export your own labeled ground truth data from the Image Labeler app by selecting Export and then To Workspace. Name the variable gTruth.
load gTruth.matExtract ROI Position and Label Data
The LabelData property of the gTruth object stores the polygon label data as a table with one column for each label.
labelData = gTruth.LabelData
labelData=1×2 table
normal tumor
____________ __________
{4×2 double} {4×1 cell}
Use the helper function gTruthtoXY, defined at the end of this example, to convert the xy-position data and labels stored in labelData into a format accepted as an input to the polyToBlockedImage function. gTruthtoXY assigns the numeric label 1 to the normal tissue ROI and the label 2 to the tumor tissue ROIs.
[roiPositions,roiLabels] = gTruthtoXY(labelData)
roiPositions=5×1 cell array
{ 4×2 double}
{14×2 double}
{16×2 double}
{20×2 double}
{12×2 double}
roiLabels = 5×1
1
2
2
2
2
Create Labeled Blocked Image
Select the desired resolution level for the new blocked image. This choice is a tradeoff between efficiency and accuracy. Using a coarser resolution level decreases processing time and storage size. Using a finer resolution level increases the level of detail preserved in the mask. You can use coarse resolution levels for regular ROIs, like polygons. For small, freehand ROIs, a fine resolution level is more appropriate. For this example, use an intermediate resolution level.
maskLevel = 2;
Specify the image size for the new blocked image to match that of the original image, bim, at the desired resolution level.
imageSize = bim.Size(maskLevel,1:2);
Create a labeled blocked image. Maintain the spatial referencing of the original blocked image, bim, at the desired resolution level. By default, polyToBlockedImage assigns pixels that do not fall inside any ROI the numeric label 0.
bLabeled = polyToBlockedImage(roiPositions,roiLabels,imageSize, ... BlockSize=bim.BlockSize(maskLevel,1:2), ... WorldStart=bim.WorldStart(maskLevel,1:2),WorldEnd=bim.WorldEnd(maskLevel,1:2));
Display the labeled blocked image overlaid on the original image. The regions corresponding to the tumor and normal tissue polygons of the groundTruth object are shown in red and green, respectively. To distinguish the normal tissue outside of the ROIs from the background using a binary mask, see Create Labeled Blocked Image from ROIs and Masks.
hbim = bigimageshow(bim); showlabels(hbim,bLabeled,Colormap=[0 0 1; 0 1 0; 1 0 0])

Supporting Function
The gTruthtoXY helper function converts the polygon ROI coordinates and label data stored in the table labelData into cell arrays suitable for input into the polyToBlockedImage function.
function [roiPositions,roiLabels] = gTruthtoXY(labelData) totalROIs = numel(labelData{1,1}) + numel(labelData{1,2}{:}); roiPositions = cell(totalROIs,1); roiLabels = zeros(totalROIs,1); % Obtain label names from the labelData table labelName = labelData.Properties.VariableNames; roiIdx = 1; % Initialize ROI index % Loop through all labels % Assign a numeric label of 2 to tumor tissue; 1 for normal tissue for j = 1:numel(labelData) % All ROIs for a given label data = labelData{1,j}{:}; if(isequal(labelName{j},"tumor")) for k = 1:numel(data) roiLabels(roiIdx) = 2; roiPositions{roiIdx} = data{k}; roiIdx = roiIdx + 1; end else % For other ROI labels roiLabels(roiIdx) = 1; roiPositions{roiIdx} = data; roiIdx = roiIdx + 1; end end end
See Also
polyToBlockedImage | blockedImage | Image
Labeler (Computer Vision Toolbox)
Topics
- Label Large Images in the Image Labeler (Computer Vision Toolbox)
- Create Labeled Blocked Image from ROIs and Masks